| |
 |
| How to use PEMPNI? |
|
| |
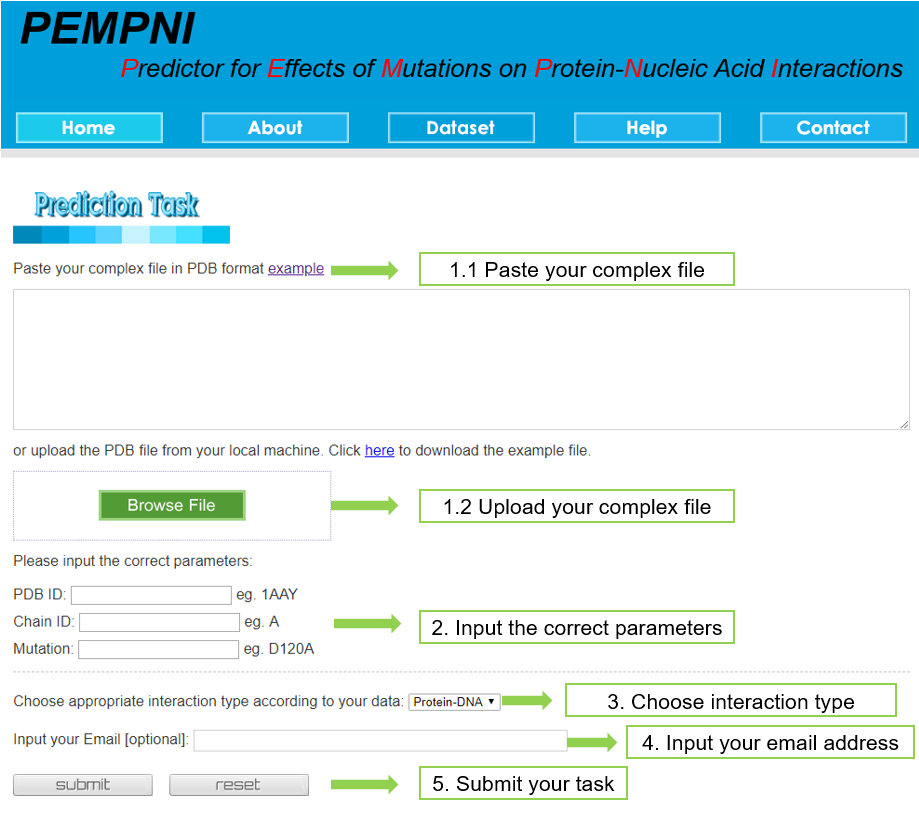
In order to use PEMPNI, you should provide a complex file in PDB format and conduct a 5-step submission as follows: (i) Paste or upload a PDB file; (ii) Input the correct parameters; (iii) Choose appropriate interaction type; (iv) Input your email address (optional); (v) Click the submit button. Users could directly submit the raw PDB file to our webserver, because a program can automatically extract the lines including atomic coordinates and remove other information.
|
| |
| Results from PEMPNI |
|
| |
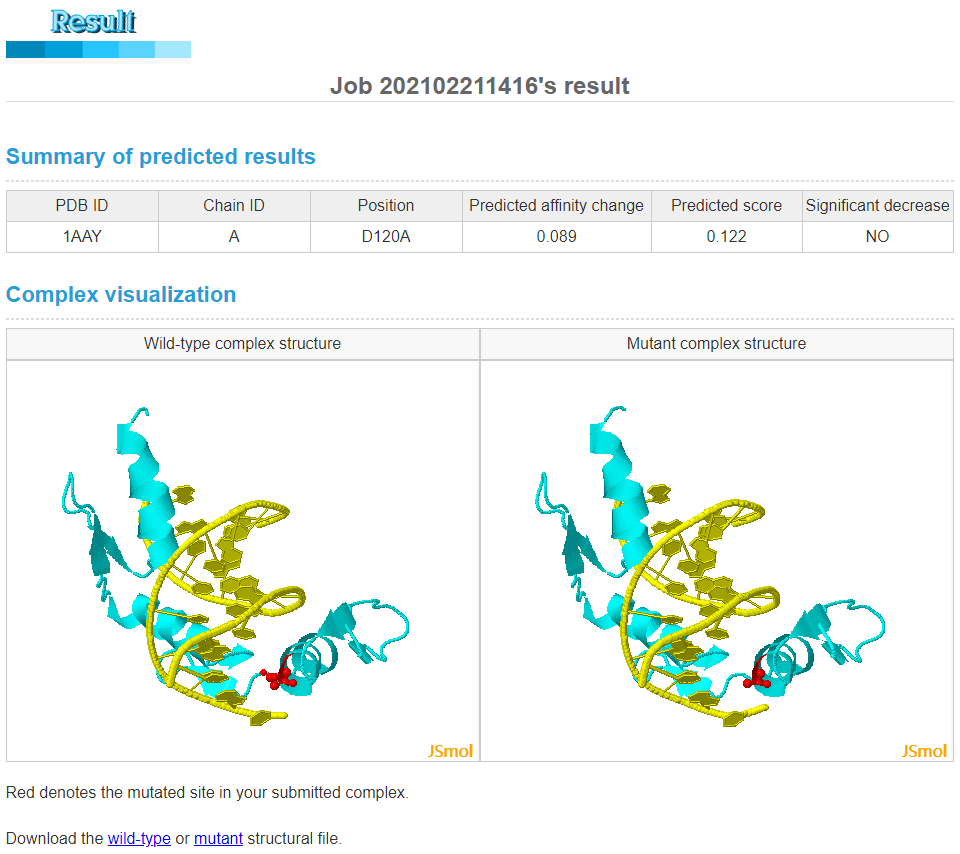
In the result page, we provide a summary of predicted results (including the predicted binding affinity change and the predicted score of significantly decreasing the affinity) and display the wild-type and mutant complex structures. You could download the structural file using the following hyperlink. Predicted affinity changes are typically in the range of -3 to 5, and positive and negative values correspond to destabilizing and stabilizing effects, respectively.Predicted scores are in the range of 0 to 1, and if this score is greater than 0.47 for MPDs or 0.32 for MPRs, the submitted mutation could significantly decrease binding affinity.
|
| |
| |
|
|
|

