| |
 |
| The binding affinities of protein-nucleic acid interactions may be altered due to missense mutations occurring in DNA- or RNA-binding proteins, therefore resulting in various diseases. Unfortunately, a systematic comparison and prediction of the effects of mutations on protein-DNA and protein-RNA interactions (these two mutation classes are termed MPDs and MPRs, respectively) is still lacking. Here, we demonstrated that these two types of mutations could generate similar or different tendencies for binding free energy changes in terms of the properties of mutated residues. We then developed regression algorithms separately for MPDs and MPRs by introducing novel geometric partition-based energy features and interface-based structural features. Through feature selection and ensemble learning, similar computational frameworks that combined energy- and nonenergy-based models were established to estimate the binding affinity changes resulting from MPDs and MPRs, but the selected features for the final models were different and therefore reflected the specificities of the two mutation classes. Furthermore, the proposed methodology was extended to the identification of mutations that significantly decreased the binding affinities. Our algorithm generally performed better than the existing methods for both the regression and classification tasks. In summary, this work provides a comprehensive survey and effective predictors for the impacts of MPDs and MPRs, which could lead to a deeper understanding of the mechanism of protein-nucleic acid interactions. |
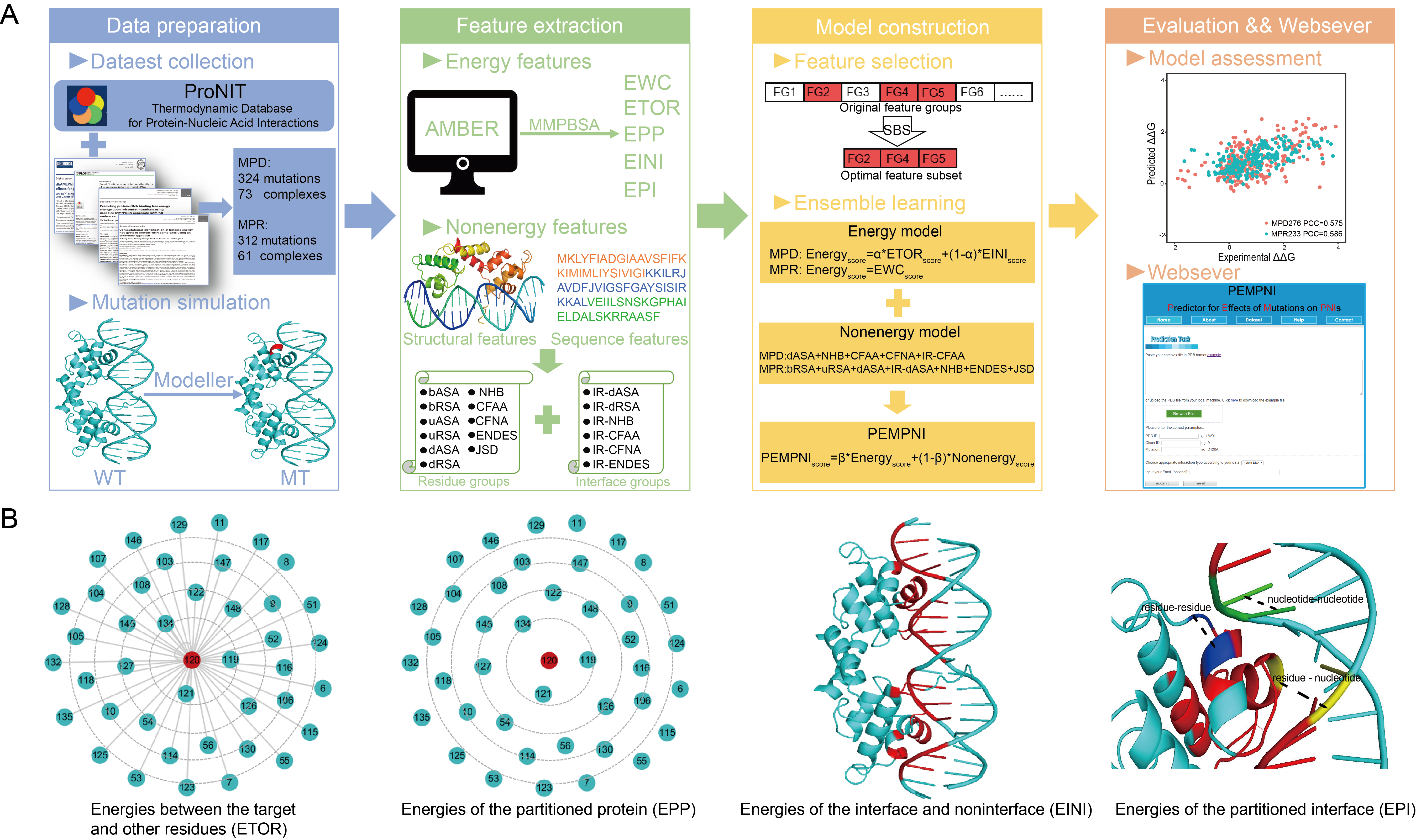
Schematic representation of PEMPNI and energy features. |
| Citation: Systematic comparison and prediction of the effects of missense mutations on protein-DNA and protein-RNA interactions. |
| |
|
|
|

