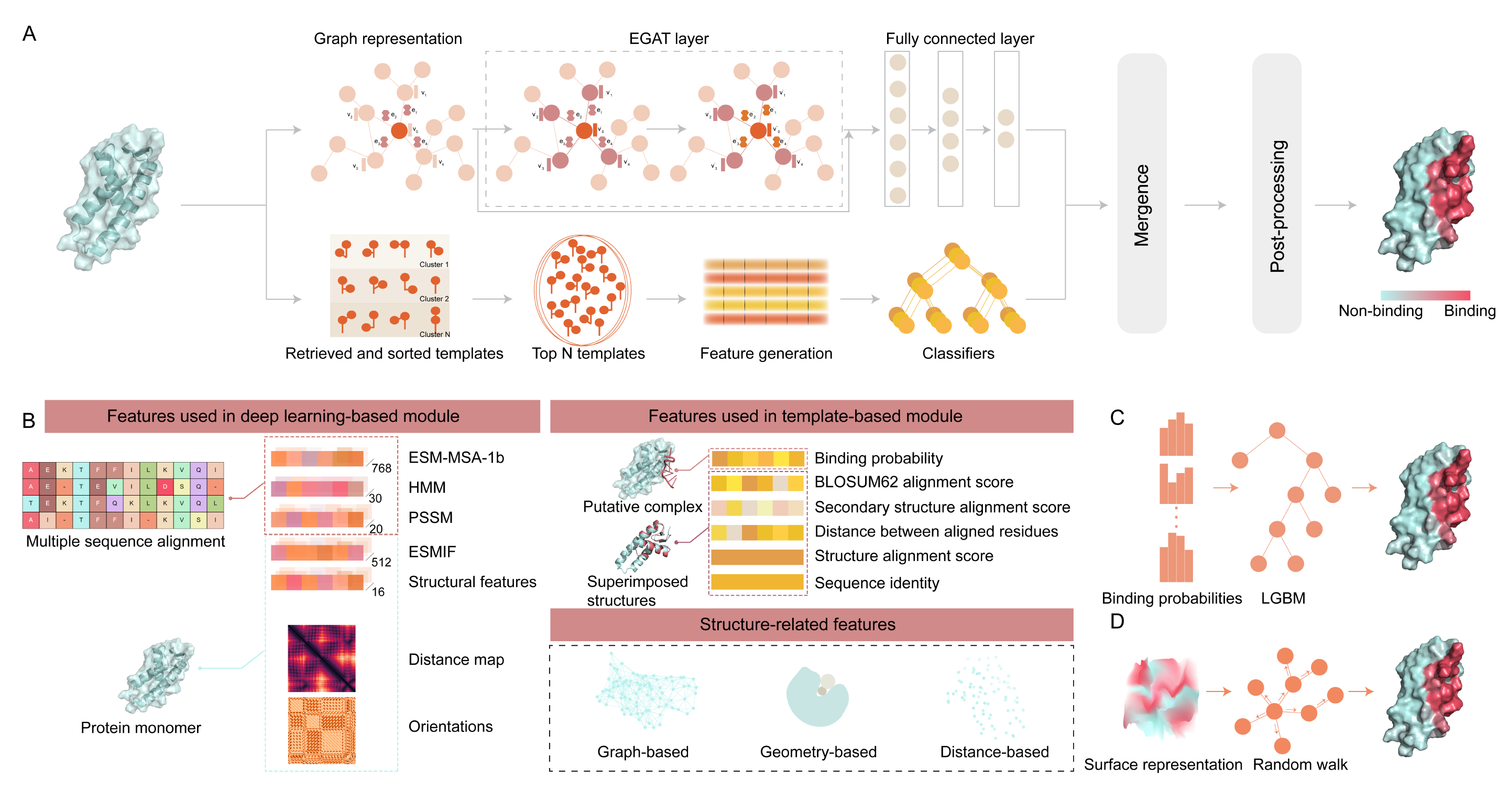
Overview
NABind is a novel structure-based method, which consists of four parts: a deep learning-based module, a template-based module, a merging module, and a post-processing module. In the deep learning phase, the protein structure is converted into a graph representation, and diversified sequence and structural descriptors are extracted and assigned to the nodes and edges of the graph. The template-based module searches the query structure against a template library, generates template-related features, and predicts the binding probabilities of residues through machine learning classifiers. The merging module integrates the outputs of the deep learning and template modules using a stacking strategy to establish an ensemble classifier. The post-processing module corrects the integrative prediction results by performing random walks on networks comprising protein surface residues.